R Practice 2
1 Tipos de datos
x <- 28 print(class(x)) y <- "R is fantastic" print(class(y)) z <- TRUE print(class(z))
[1] "numeric" [1] "character" [1] "logical"
1.1 Vectores
datos <- c(2,1,3,-1,-10,0,0,1)
print(datos)
[1] 2 1 3 -1 -10 0 0 1
1.1.1 Number sequences
print(1:10) print(seq(10)) print(seq(1, 10, by=3)) print(rep(1:4, 2))
[1] 1 2 3 4 5 6 7 8 9 10 [1] 1 2 3 4 5 6 7 8 9 10 [1] 1 4 7 10 [1] 1 2 3 4 1 2 3 4
1.1.2 Access
print(datos)
print(datos[1])
print(datos[-4])
print(datos[c(1,3,5)])
print(datos[3:5])
print(v <- datos>1)
print(datos[v])
[1] 2 1 3 -1 -10 0 0 1 [1] 2 [1] 2 1 3 -10 0 0 1 [1] 2 3 -10 [1] 3 -1 -10 [1] TRUE FALSE TRUE FALSE FALSE FALSE FALSE FALSE [1] 2 3
1.1.3 Index names
Vectors can have names on their indexes.
v = seq(1:5) names(v) <- c("Lun", "Mar", "Mie", "Jue", "Vie") print(v) print(names(v))
Lun Mar Mie Jue Vie 1 2 3 4 5 [1] "Lun" "Mar" "Mie" "Jue" "Vie"
1.1.4 Fncs.
If a function does not return anything, then the vector may have a NA value.
print(length(datos)) print(min(datos)) print(max(datos)) print(sum(datos)) print(mean(datos)) print(median(datos)) print(sort(datos)) print(unique(datos)) print(which(datos > 1)) print(which.max(datos)) print(which.min(datos))
[1] 8 [1] -10 [1] 3 [1] -4 [1] -0.5 [1] 0.5 [1] -10 -1 0 0 1 1 2 3 [1] 2 1 3 -1 -10 0 [1] 1 3 [1] 3 [1] 5
plot(datos)

1.1.5 Factor
Nominal (unordered) or categorical (ordered) factor.
Used on Likert scale.
day_vector <- c('evening', 'morning', 'afrernoon', 'midday', 'midnight', 'evening') factor_day <- factor(day_vector, order=TRUE, levels=c('morning', 'midday', 'afternoon', 'evening', 'midnight'))
color_vector <- c('blue', 'red', 'yellow', 'green', 'white') factor_color <- factor(color_vector) print(factor_color)
[1] blue red yellow green white Levels: blue green red white yellow
1.2 Data Frame
A new data frame can be created by using data.frame function.
a <- c(10, 20, 30 , 40) b <- c('book', 'pen', 'textbook', 'pencil_case') c <- c(TRUE, FALSE, TRUE, FALSE) d <- c(2.5, 8, 10, 7) df <- data.frame(a, b, c, d) print(df)
a b c d 1 10 book TRUE 2.5 2 20 pen FALSE 8.0 3 30 textbook TRUE 10.0 4 40 pencil_case FALSE 7.0
Change names on the data frame.
names(df) <- c("ID", "items", "store", "price") print(df)
ID items store price 1 10 book TRUE 2.5 2 20 pen FALSE 8.0 3 30 textbook TRUE 10.0 4 40 pencil_case FALSE 7.0
Print the structure and description of the data frame.
str(df)
'data.frame': 4 obs. of 4 variables: $ ID : num 10 20 30 40 $ items: Factor w/ 4 levels "book","pen","pencil_case",..: 1 2 4 3 $ store: logi TRUE FALSE TRUE FALSE $ price: num 2.5 8 10 7
1.2.1 Slices
print(df[1,2])
print(df[1:2,])
print(df[1:3, 3:4])
print(df[,1])
print(df[,"ID"])
[1] book Levels: book pen pencil_case textbook ID items store price 1 10 book TRUE 2.5 2 20 pen FALSE 8.0 store price 1 TRUE 2.5 2 FALSE 8.0 3 TRUE 10.0 [1] 10 20 30 40 [1] 10 20 30 40
1.2.2 Add columns
The amount of data in the vector must be of the same length of the data frame's columns.
quantity <- c(10, 35, 40, 5) df$quantity <- quantity print(df)
ID items store price quantity 1 10 book TRUE 2.5 10 2 20 pen FALSE 8.0 35 3 30 textbook TRUE 10.0 40 4 40 pencil_case FALSE 7.0 5
1.2.3 Subset
print(subset(df, subset=price > 5))
ID items store price quantity 2 20 pen FALSE 8 35 3 30 textbook TRUE 10 40 4 40 pencil_case FALSE 7 5
1.2.4 Merge - Joins
Full merge is when both columns has data. Partial merge is when one of the column has data that is not in the other, then a NA value will be assigned.
x <- c("n1", "n2") y <- c(1, 2) z <- data.frame(x, y) print(z) t <- c("a", "b") s <- c(1, 2) o <- data.frame(t, s) print(o) q <- merge(z, o, by.x = "y", by.y = "s") print(q)
x y 1 n1 1 2 n2 2 t s 1 a 1 2 b 2 y x t 1 1 n1 a 2 2 n2 b
Partial match. The following snippet adds a new row into z.
add_z <- c("n2", 3) z <- rbind(z, add_z) print(z) # Partial match q <- merge(z, o, by.x = "y", by.y = "s", all.x = TRUE) print(q) # Total match q <- merge(z, o, by.x = "y", by.y = "s") print(q)
x y 1 n1 1 2 n2 2 3 n2 3 y x t 1 1 n1 a 2 2 n2 b 3 3 n2 <NA> y x t 1 1 n1 a 2 2 n2 b
2 Plotting
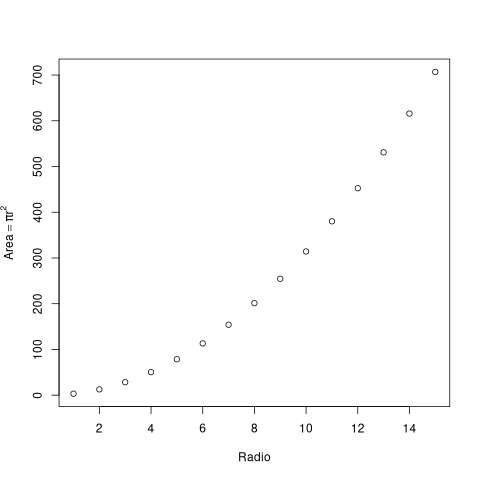
x <- 1:15 y <- pi*x^2 print(x) print(y)
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 [1] 3.141593 12.566371 28.274334 50.265482 78.539816 113.097336 [7] 153.938040 201.061930 254.469005 314.159265 380.132711 452.389342 [13] 530.929158 615.752160 706.858347
plot(x, y, xlab="Radio", ylab=expression(Area == pi*r^2))
Correlation. If p-value < 0.05, then they are correlated. If cor is near 1, the more correlated they are.
res <- cor.test(x,y)
print(res)
Pearson's product-moment correlation
data: x and y
t = 15.029, df = 13, p-value = 1.348e-09
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.9168624 0.9910175
sample estimates:
cor
0.9724093
celsius <- -25:30 fahrenheit <- 9/5*celsius+32 x <- data.frame(Celsius=celsius, Fahrenheit=fahrenheit) plot(x)

plot(celsius~fahrenheit)
2.1 Bar plot
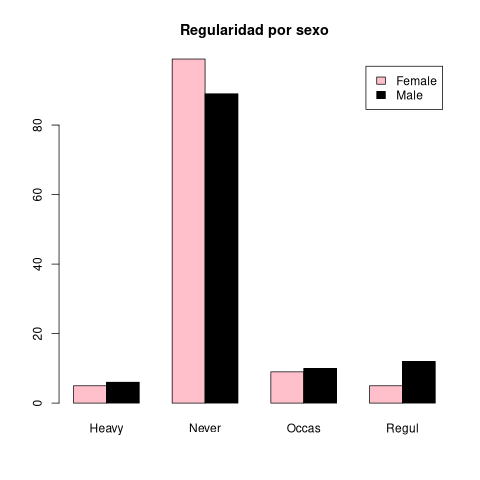
library(MASS) t <- table(survey$Sex, survey$Smoke) print(t)
Heavy Never Occas Regul
Female 5 99 9 5
Male 6 89 10 12
barplot(table(survey$Sex, survey$Smoke), beside=TRUE)

barplot(t, beside=TRUE)

barplot(t, main="Regularidad por sexo", legend.text=c("Female", "Male"), beside=TRUE, col = c("pink", "black"))
2.2 Pie plot
f <- table(survey$Sex)
print(f)
Female Male 118 118
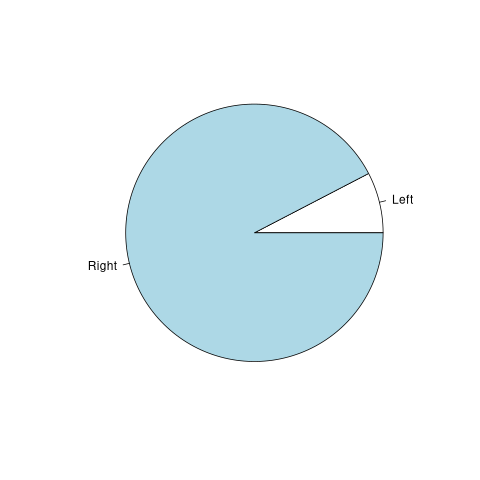
f <- table(survey$W.Hnd)
print(f)
Left Right 18 218
pie(f)
z <- round(f/sum(f)*100) print(z) lbs <- paste(names(f), z, "%", sep=" ") print(lbs)
Left Right
8 92
[1] "Left 8 %" "Right 92 %"
pie(f, main="Cantidad de encuestados diestros y zurdos", labels=lbs)
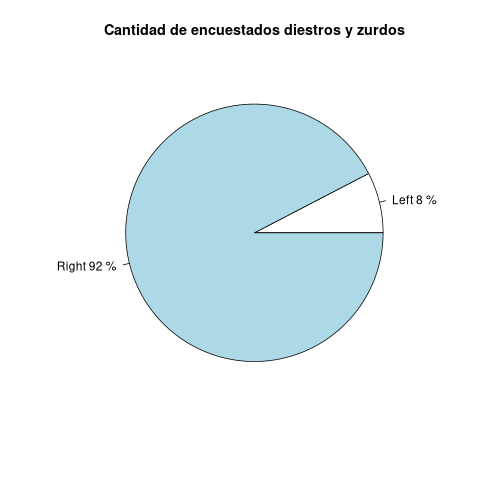
pie(f, main="Cantidad de encuestados diestros y zurdos", labels=lbs,
col=rainbow(length(lbs)))

2.3 Histograms
hist(survey$Age)

This is a normal distributed histogram.
hist(survey$Height)

Testing normality.
print(shapiro.test(survey$Age))
Shapiro-Wilk normality test data: survey$Age W = 0.45642, p-value < 2.2e-16
If p-value > 0.05 then it is normal.
print(shapiro.test(survey$Height))
Shapiro-Wilk normality test data: survey$Height W = 0.98841, p-value = 0.08844
2.3.1 Example
#........................................Histograma tiempo <- c(11.50 , 10.26, 10.08, 13.00, 11.14, 13.73, 13.41, 10.44, 11.36, 14.40, 11.64, 12.39, 12.82, 14.25, 15.41, 14.35, 9.35, 12.40, 9.04, 15.30, 14.79, 15.27, 10.63, 14.30, 15.48, 14.80, 8.78, 14.00, 13.09, 10.00, 12.20, 11.70, 15.37, 11.81, 10.06, 12.49, 8.58, 11.32, 12.20, 12.45, 11.28, 12.60, 14.36, 13.08, 13.50, 12.68, 9.19, 14.32, 12.17, 9.10) sum <- min(tiempo) for (i in 1:7) { sum <- sum + 0.99 print(sum) } #........................................Barplot x <- c(rep("muy de acuerdo",10), rep("de acuerdo", 10), rep("poco de acuerdo", 10), rep("para nada de acuerdo", 10)) factor_x <- factor(x, order=TRUE, levels = c("para nada de acuerdo", "poco de acuerdo", "de acuerdo", "muy de acuerdo")) w <- as.numeric(factor_x) y <- c(rep("hombre",18), rep("mujer",2), rep("mujer",18), rep("hombre",2)) #........................................Boxplot g <- c(36,25,37,24,39,20,36,45,31,31,39,24,29,23,41,40,33,24,34,40) g
[1] 9.57 [1] 10.56 [1] 11.55 [1] 12.54 [1] 13.53 [1] 14.52 [1] 15.51 [1] 36 25 37 24 39 20 36 45 31 31 39 24 29 23 41 40 33 24 34 40
tiempo <- c(11.50 , 10.26, 10.08, 13.00, 11.14, 13.73, 13.41, 10.44, 11.36, 14.40, 11.64, 12.39, 12.82, 14.25, 15.41, 14.35, 9.35, 12.40, 9.04, 15.30, 14.79, 15.27, 10.63, 14.30, 15.48, 14.80, 8.78, 14.00, 13.09, 10.00, 12.20, 11.70, 15.37, 11.81, 10.06, 12.49, 8.58, 11.32, 12.20, 12.45, 11.28, 12.60, 14.36, 13.08, 13.50, 12.68, 9.19, 14.32, 12.17, 9.10)
| 11.5 |
| 10.26 |
| 10.08 |
| 13 |
| 11.14 |
| 13.73 |
| 13.41 |
| 10.44 |
| 11.36 |
| 14.4 |
| 11.64 |
| 12.39 |
| 12.82 |
| 14.25 |
| 15.41 |
| 14.35 |
| 9.35 |
| 12.4 |
| 9.04 |
| 15.3 |
| 14.79 |
| 15.27 |
| 10.63 |
| 14.3 |
| 15.48 |
| 14.8 |
| 8.78 |
| 14 |
| 13.09 |
| 10 |
| 12.2 |
| 11.7 |
| 15.37 |
| 11.81 |
| 10.06 |
| 12.49 |
| 8.58 |
| 11.32 |
| 12.2 |
| 12.45 |
| 11.28 |
| 12.6 |
| 14.36 |
| 13.08 |
| 13.5 |
| 12.68 |
| 9.19 |
| 14.32 |
| 12.17 |
| 9.1 |
trange <- range(tiempo) tmax <- max(tiempo) tmin <- min(tiempo) print(trange) trange2 <- tmax-tmin print(trange2)
[1] 8.58 15.48 [1] 6.9
int_clase <- sqrt(length(tiempo)) print(int_clase) int_clase <- round(int_clase) print(int_clase)
[1] 7.071068 [1] 7
amplitud <- trange2/int_clase print(amplitud) amplitud <- round(amplitud) print(amplitud)
[1] 0.9857143 [1] 1
The number of classes in a histogram can be changed with the nclass parameter.
hist(survey$Height, nclass=20)
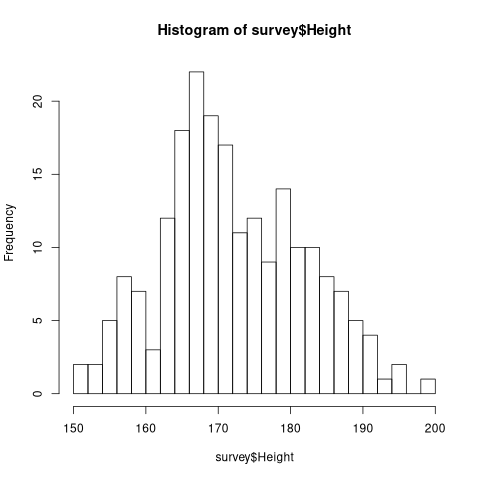
2.3.2 Acumulative (Frequent) Histograms
z$counts has the frequency of the elements in the histograms.
z <- hist(survey$Height)
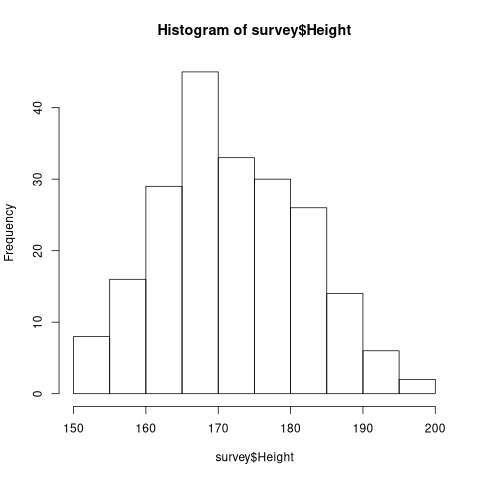
print(z$counts)
| 8 |
| 16 |
| 29 |
| 45 |
| 33 |
| 30 |
| 26 |
| 14 |
| 6 |
| 2 |
z$counts <- cumsum(z$counts)
print(z$counts)
| 8 |
| 24 |
| 53 |
| 98 |
| 131 |
| 161 |
| 187 |
| 201 |
| 207 |
| 209 |
plot(z)
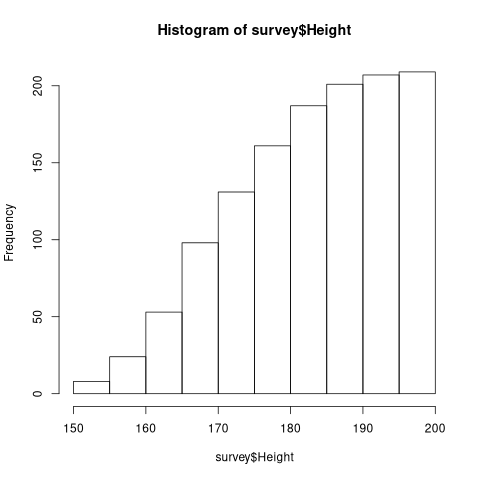
2.4 Box Plot
g <- c(36,25,37,24,39,20,36,45,31,31,39,24,29,23,41,40,33,24,34,40) bp1 <- boxplot(g)

print(bp1)
$stats
[,1]
[1,] 20.0
[2,] 24.5
[3,] 33.5
[4,] 39.0
[5,] 45.0
$n
[1] 20
$conf
[,1]
[1,] 28.37717
[2,] 38.62283
$out
numeric(0)
$group
numeric(0)
$names
[1] "1"
horizontal plot
bp2 <- boxplot(g, horizontal=TRUE)
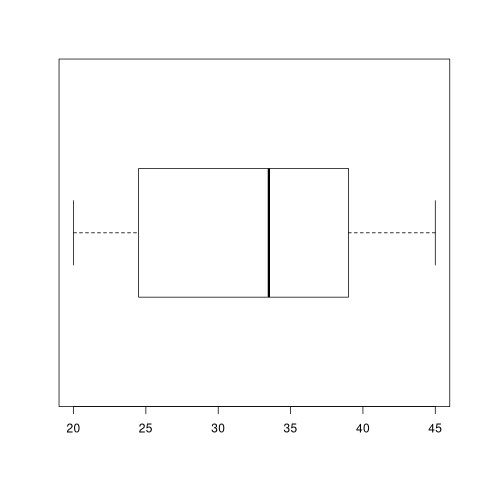
print(bp2)
$stats
[,1]
[1,] 20.0
[2,] 24.5
[3,] 33.5
[4,] 39.0
[5,] 45.0
$n
[1] 20
$conf
[,1]
[1,] 28.37717
[2,] 38.62283
$out
numeric(0)
$group
numeric(0)
$names
[1] "1"
bp1$stats returns the quartiles values.
print(sort(g)) print(bp1$stats)
[1] 20 23 24 24 24 25 29 31 31 33 34 36 36 37 39 39 40 40 41 45
[,1]
[1,] 20.0
[2,] 24.5
[3,] 33.5
[4,] 39.0
[5,] 45.0
2.4.1 Quartiles
g1 <- sort(g)
print(g1)
[1] 20 23 24 24 24 25 29 31 31 33 34 36 36 37 39 39 40 40 41 45
First quartile is the arithmetic media between the 4th quarter and the next values.
print( (g1[length(g1)/4] + g1[length(g1)/4 + 1] ) / 2 )
[1] 24.5
print(median(g1))
[1] 33.5
3 Correlation
print(min(survey$NW.Hnd, na.rm=TRUE))
[1] 12.5
plot(survey$Wr.Hnd, survey$NW.Hnd)

res <- cor.test(survey$Wr.Hnd, survey$NW.Hnd)
print(res)
Pearson's product-moment correlation
data: survey$Wr.Hnd and survey$NW.Hnd
t = 45.712, df = 234, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.9336780 0.9597816
sample estimates:
cor
0.9483103
To ensure that the Pearson method is applied, use the method parameter.
res <- cor.test(survey$Wr.Hnd, survey$NW.Hnd, method="pearson") print(res)
Pearson's product-moment correlation
data: survey$Wr.Hnd and survey$NW.Hnd
t = 45.712, df = 234, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.9336780 0.9597816
sample estimates:
cor
0.9483103
3.1 Wilcox correlation
Some comparison between a Likert (nominal) with another factor it has to be applied with the corresponding parametric or non-parametric correlation.
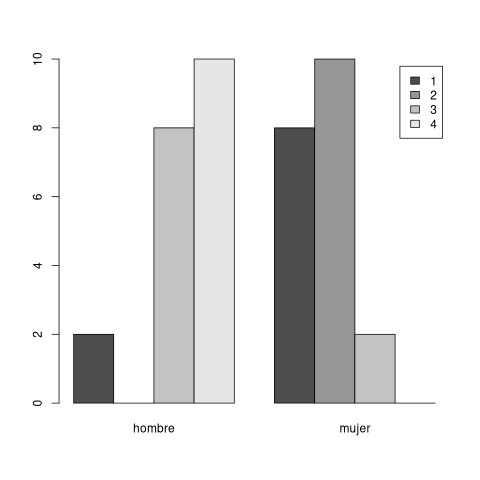
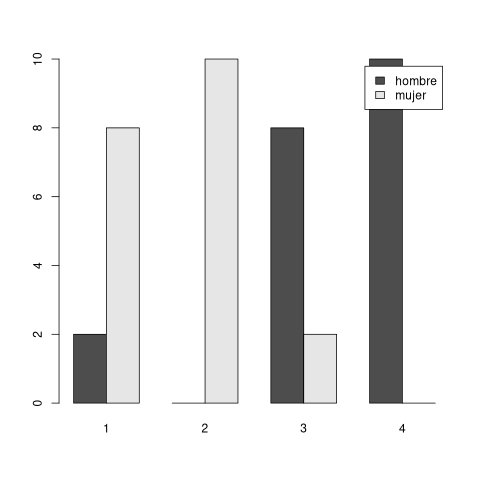
x <- c(rep("muy de acuerdo",10), rep("de acuerdo", 10), rep("poco de acuerdo", 10), rep("para nada de acuerdo", 10)) factor_x <- factor(x, order=TRUE, levels = c("para nada de acuerdo", "poco de acuerdo", "de acuerdo", "muy de acuerdo")) print(factor_x) w <- as.numeric(factor_x) print(w) y <- c(rep("hombre",18), rep("mujer",2), rep("mujer",18), rep("hombre",2)) print(y)
[1] muy de acuerdo muy de acuerdo muy de acuerdo [4] muy de acuerdo muy de acuerdo muy de acuerdo [7] muy de acuerdo muy de acuerdo muy de acuerdo [10] muy de acuerdo de acuerdo de acuerdo [13] de acuerdo de acuerdo de acuerdo [16] de acuerdo de acuerdo de acuerdo [19] de acuerdo de acuerdo poco de acuerdo [22] poco de acuerdo poco de acuerdo poco de acuerdo [25] poco de acuerdo poco de acuerdo poco de acuerdo [28] poco de acuerdo poco de acuerdo poco de acuerdo [31] para nada de acuerdo para nada de acuerdo para nada de acuerdo [34] para nada de acuerdo para nada de acuerdo para nada de acuerdo [37] para nada de acuerdo para nada de acuerdo para nada de acuerdo [40] para nada de acuerdo 4 Levels: para nada de acuerdo < poco de acuerdo < ... < muy de acuerdo [1] 4 4 4 4 4 4 4 4 4 4 3 3 3 3 3 3 3 3 3 3 2 2 2 2 2 2 2 2 2 2 1 1 1 1 1 1 1 1 [39] 1 1 [1] "hombre" "hombre" "hombre" "hombre" "hombre" "hombre" "hombre" "hombre" [9] "hombre" "hombre" "hombre" "hombre" "hombre" "hombre" "hombre" "hombre" [17] "hombre" "hombre" "mujer" "mujer" "mujer" "mujer" "mujer" "mujer" [25] "mujer" "mujer" "mujer" "mujer" "mujer" "mujer" "mujer" "mujer" [33] "mujer" "mujer" "mujer" "mujer" "mujer" "mujer" "hombre" "hombre"
The Wilcoxon test has a p-value less than 0.05 which means that there is correlation between the Likert data (w or x) that is influenced by the sex (y data).
m <- data.frame(w, y) wilcox.test(w~y, alternative="two.sided", data=m) print(m)
Wilcoxon rank sum test with continuity correction data: w by y W = 360, p-value = 8.405e-06 alternative hypothesis: true location shift is not equal to 0 Warning message: In wilcox.test.default(x = c(4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 3, 3, : cannot compute exact p-value with ties w y 1 4 hombre 2 4 hombre 3 4 hombre 4 4 hombre 5 4 hombre 6 4 hombre 7 4 hombre 8 4 hombre 9 4 hombre 10 4 hombre 11 3 hombre 12 3 hombre 13 3 hombre 14 3 hombre 15 3 hombre 16 3 hombre 17 3 hombre 18 3 hombre 19 3 mujer 20 3 mujer 21 2 mujer 22 2 mujer 23 2 mujer 24 2 mujer 25 2 mujer 26 2 mujer 27 2 mujer 28 2 mujer 29 2 mujer 30 2 mujer 31 1 mujer 32 1 mujer 33 1 mujer 34 1 mujer 35 1 mujer 36 1 mujer 37 1 mujer 38 1 mujer 39 1 hombre 40 1 hombre
barplot(table(m), beside=TRUE, legend=TRUE)
barplot(table(y, w), beside=TRUE, legend=TRUE)
4 License of This Work
This work is licensed under the Creative Commons Attribution-NoDerivatives 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nd/4.0/.

R Practice 2 by Gimenez Christian is licensed under a Creative Commons Attribution-NoDerivatives 4.0 International License.